major update August 2025 - previous version (SPSS via virtual computing, R/RStudio local system install without docker) is here
It can be difficult to manage everybody's individual technology issues, so it's important to make sure we all have similar set ups. Pick a location on your computer's file system, and create a new folder called psychstats596 - leave it empty for now.
R is a statistical programming language. RStudio is an interactive development environment (IDE) built to make it easier to run, view, and document your work that uses the R language. R and Rstudio are open-source, so you can install them on your own.
if you already have R and Rstudio installed, make sure you have an R version 4 or greater (e.g., 4.0.4 or higher version number), and RStudio 1.4 or greater. Class materials are tested in R version 4.3.3 and RStudio version 2023.12.1+402, with tidyverse version 2.0.0
getRversion() in the RStudio consoleIf you already have R and RStudio installed, I still recommend running it from a docker container (see below). But if you are happy to manage package versions yourself and troubleshoot any conflicts that arise then I won't stop you.
One of the biggest challenges in creating reproducable research is that any work you do on a computer (e.g., design, simulation, analysis) relies on the programs and environment that are particular to your computer. This means it is not always easy for someone to recreate your work A Docker container basically simulates a whole computing environment on your host computer. In this case I have created a Docker image for this class that you can download and run on your own computer. This image can be used to run a container that has the RStudio environment and most of the pckages you will need for class (I left some packages uninstalled so that you will have some experience installing packages).
Steps to get a container running:
Now, you can pull the Docker image for this class and "spin up a container" based on the image. The image is called stored on hub.docker.com (the most common location for sharing docker images).
jamilfelipe/psych596applearm64-rstudio_app. (to check if you have an arm64 processor, open a terminal and type uname -m if the output is arm64 then it is, otherwise it is an amd64 type chip.jamilfelipe/psych596amd64-rstudio_app8787 in the "Host port" text box. This will designate a port number that you will use to access the running container through a web browser (if you have multiple containers running , you will need to use a different port numbers for each)./home/rstudio/psychstats596. Click "Run".localhost:8787 -- when the username/password box comes up enter rstudio for the user, and paste in the password that you copied. You should see the RStudio environment come up in the browser.Notes on using containers:
quit() in the console and then click "Start New Session"psychstats596 folder) -- see this guide for information about writing a new image with the changes you made in the containerFollow instructions here, start at #2
This video (linked in the Syllabus also) describes the workflow that we will use in class. These are the basic steps in the workflow:
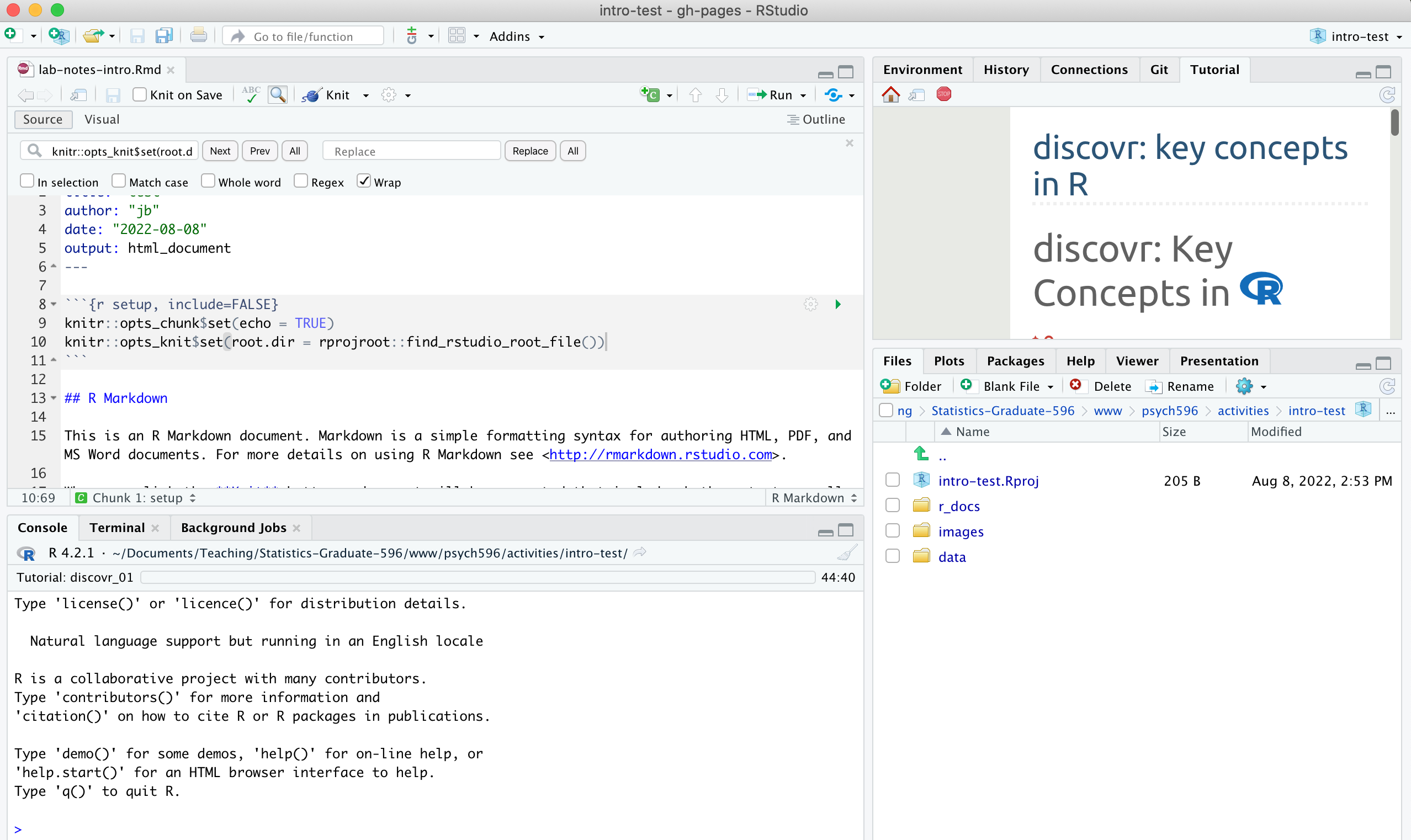
1. Create a folder containing an RStudio project (*.Rproj file) for the lab activity each week. This week, make a folder called "intro-essentials" and then use File -> New Project -> Existing Folder to create an R project file in the "intro-essentials" folder. Open the project in your current session.
2. Inside the folder you made for the project, create new folders called "data", "r_docs", and "images". You can create the folders through the Files tab in the lower right RStudio Pane, or as you normally would in Windows or MacOS.
3. Create an R Markdown file called "lab-notes-intro" and save it in "r_docs" folder. Use File ->New File -> R Markdown... then Save (on the RStudio menu). The markdown file will open in the top left RStudio pane - this is where you will write your R code and where you will take notes. When you reach a point where you want to share the document you can use the Knit option to generate a report containing your code, notes, and visualizations.
4. Delete the template text starting from "## R Markdown" down to the end of the file.
5. For future activities, write your code inside code "chunks", and run chunks in order when writing/testing code. When you want to generate a report (e.g., an html file that you can share), use the Knit button.
- the start of a chunk is designated by a line that starts with 3 backticks ` followed by {r chunk-name}. The end of a chunk is designated by a line with 3 backticks.
- in the "setup" code chunk, add this line to set the working directory (see here for explanation):
knitr::opts_knit$set(root.dir = rprojroot::find_rstudio_root_file())
6. Write your notes above or below code chunks. Characters like # and * are used for markdown-style formatting of the report as described in this pdf.
7. For this week, just write "I created an R Markdown document" as a note in your R Markdown file, then save it and submit the *.Rmd file in the Canvas lab activity assignment.
When you have set up your project, your RStudio environment should look something like this:

Run this in the Console: learnr::initialize_tutorial()
Go to the Tutorial Pane (top right) and click "start tutorial" for any tutorial that interests you. it will take a minute or two to load - use the "pop-out" button  to open the tutorial in a larger view.
to open the tutorial in a larger view.